Confronting data sparsity to identify potential sources of Zika virus spillover infection among primates
Barbara Han
Subhabrata Majumdar
Flavio Calmon
Benjamin Glicksberg
Raya Horesh
Abhishek Kumar
Elisa von Marschall
Dennis Wei
Aleksandra Mojsilović
Kush Varshney
Published at
Epidemics
2019
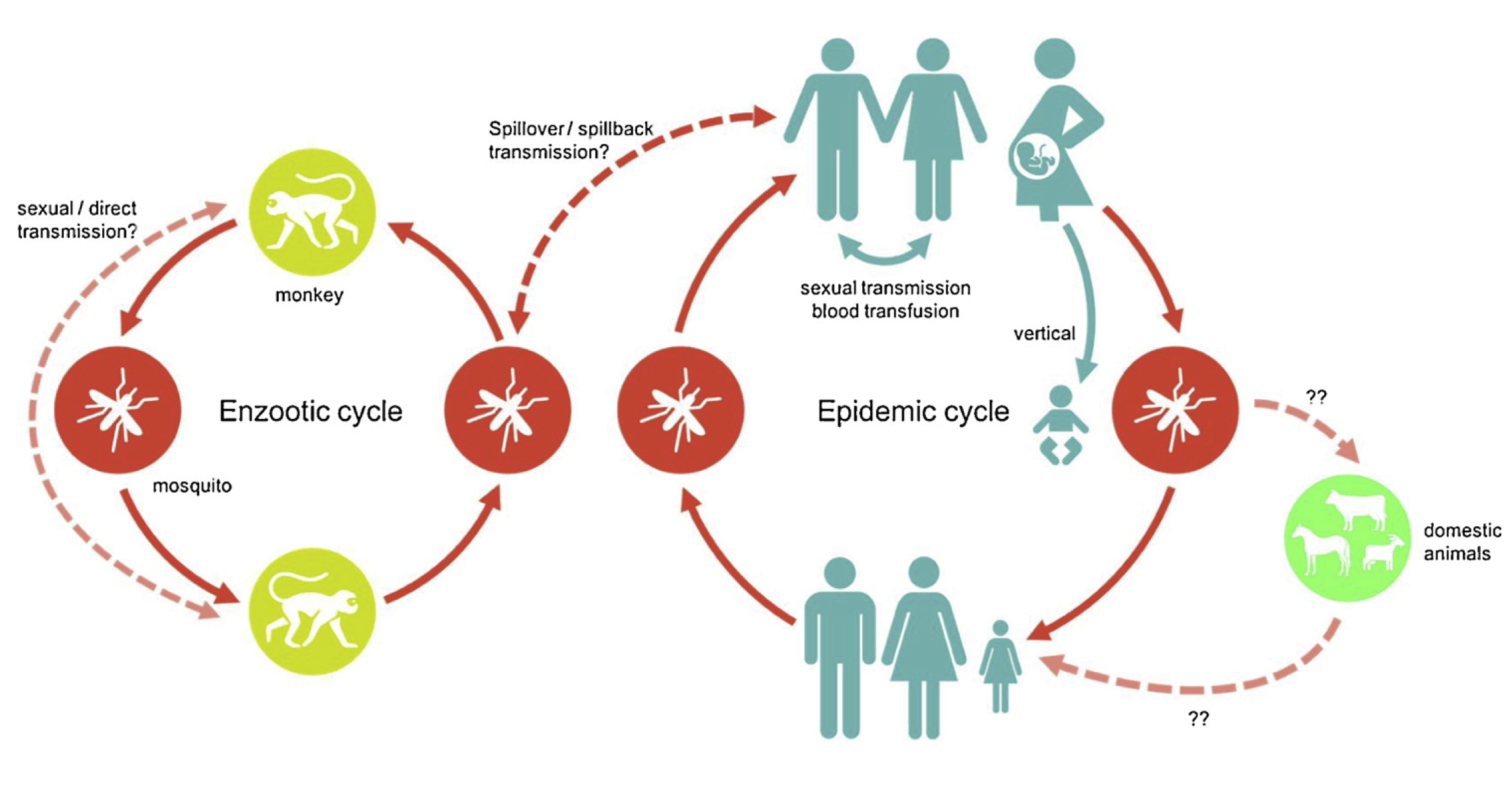
Abstract
The recent Zika virus (ZIKV) epidemic in the Americas ranks among the largest
outbreaks in modern times. Like other mosquito-borne flaviviruses, ZIKV
circulates in sylvatic cycles among primates that can serve as reservoirs of
spillover infection to humans. Identifying sylvatic reservoirs is critical to
mitigating spillover risk, but relevant surveillance and biological data remain
limited for this and most other zoonoses. We confronted this data sparsity by
combining a machine learning method, Bayesian multi-label learning, with a
multiple imputation method on primate traits. The resulting models distinguished
flavivirus-positive primates with 82% accuracy and suggest that species posing
the greatest spillover risk are also among the best adapted to human
habitations. Given pervasive data sparsity describing animal hosts, and the
virtual guarantee of data sparsity in scenarios involving novel or emerging
zoonoses, we show that computational methods can be useful in extracting
actionable inference from available data to support improved epidemiological
response and prevention.